More Information
Submitted: September 09, 2022 | Approved: September 23, 2022 | Published: September 24, 2022
How to cite this article: Muluneh A, Woldemariyam FT. Determination of foot-and-mouth disease serotypes from naturally infected cattle by solid phase competitive ELISA (SPCE) techniques. Ann Biomed Sci Eng. 2022; 6: 014-018.
DOI: 10.29328/journal.abse.1001017
Copyright License: © 2022 Muluneh A, et al. This is an open access article distributed under the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.
Keywords: FMD; Serum; Serotype solid phase competitive ELISA; Ethiopia
Determination of foot-and-mouth disease serotypes from naturally infected cattle by solid phase competitive ELISA (SPCE) techniques
Ayelech Muluneh1 and Fanos Tadesse Woldemariyam2*
1National Animal Health Institute, P.O.Box 4, Sebeta, Ethiopia
2Department of Biomedical Sciences, Addis Ababa University, College of Veterinary Medicine, P.O.Box, 34, Bishoftu, Ethiopia
*Address for Correspondence: Fanos Tadesse Woldemariyam, Department of Biomedical sciences, Addis Ababa University, College of Veterinary Medicine, P.O.Box, 34, Bishoftu, Ethiopia, FTW, Email: [email protected]
Objective: Foot and mouth disease (FMD) is a highly infectious and economically important disease affecting cloven-hoofed domestic and wild animals. Early diagnosis and serotyping of the agent are very important to effectively design and implement the control approach. This study was conducted on serum samples collected from Amhara, Tigray, Oromia and Addis Ababa between October 2018 to February 2020. The animals were kept under a semi-intensive to an extensive system of rearing. Serum samples with low OD values (positive) using competition NSP-ELISA were subjected to serotyping ELISA.
Results: In the present study, three serotypes were identified from 186 NSP ELISA positive sera of which 156 serotype O, 40 serotypes A and 28 serotype SAT2. In this analysis, multiple serotype infection was observed which is why the number of serotypes was beyond the samples analyzed. Among 23 samples from Addis Ababa 10, 3 and 5 were O, A and SAT2 serotypes respectively, while in samples from the Oromia region 12 were O and 3 were SAT2 serotypes. From the Amhara region, 99 samples analyzed were found to be serotype O and SAT2 in 7 of the serum samples. From the Tigray region, 30 samples were seen to have Serotype O infection, whereas 13 of them were SAT2. The proportion of serotypes identified based on the production system practices was also found that semi-intensive production takes the largest share in all three serotypes followed by extensive production. Generally, early determination of the serotype from past infection helps to aware of the epidemiology as well as the infection immunity of the herd/individual animals.
Foot-and-mouth disease is a highly infectious economically important disease affecting cloven-hoofed domestic and wild animals. It is caused by Foot and mouth diseases virus of the genus Apthovirus, family picornaviridae. This virus has seven immunologically distinct serotypes namely O, A, SAT1, 2, 3, Asia 1 and C. These serotypes again have sub-groups in their geographic presence as topotypes. So far there are over 60 topotypes [1,2].
As these serotypes or topotypes have the distinct immunological appearance, infection with one of them does not guarantee future protection. Similarly, vaccination also works within the serotype limit. Multiple infections with one or more serotypes at the same time can also happen [3,4]. In most of sub-Saharan Africa, the disease is endemic with five serotypes (O, A, SAT1, SAT2, SAT3) known to circulate [5]. Foot-and-mouth disease outbreak in Ethiopia occurs an average of 35 times a year. Most outbreaks occur from October to March in the dry period of the year. This outbreak number is underestimated because of a lack of proper reporting. Even the reported outbreaks are communicated to the responsible body but no control or prevention measures are in place [6].
Outbreak investigation and characterization using viral isolation and molecular technique is a crucial step in identifying the serotype responsible for the outbreak [7]. Serological survey methods are also very important to identify whether the animal had previous exposure or not. The most commonly used method is the non-structural protein-based detection of infected animals by discriminating from vaccine-induced antibody response. This method of diagnosis is the best method and helps to determine the prevalence of the diseases based on previous exposure but not the current status and the serotype. Serotyping of FMDV from either the serum or the isolated virus can be done by LPBE, SPCE and Virus neutralization methods. Virus neutralization also works by titrating a known antibody or antigen against a known antibody or antigen on a cell monolayer. In both cases inhibition of the cytopathic effect as a result of the presence of protective or matched antibody is indicative of the specific serotype. Solid phase blocking ELISA is a method by which serotyping of a specific virus from the serum of actively diseased animals or previously infected animals was done. This is the best method that avoids all the hurdles of virus isolation, molecular characterization, and Virus neutralization (VN) methods which all needs a high confinement facility.
Therefore the aim of this study was to serotype NSP ELISA positive serum samples using SPCE from previously identified naturally infected animals.
Study area and sampling strategy
This study was conducted on serum samples collected from Amhara, Tigray, Oromia and Addis Ababa regions between October 2018 to February 2020. The production system in which the animals were kept was from semi-intensive to extensive system. Serum samples with high OD values Using NSP-ELISA were subjected to stereotyping ELISA. Regions, Zones and districts were selected purposively and individual farms/herds and animals were selected based on systematic random sampling. Serum from cattle, swine and goats was used in the analysis. Animals below six months of age were not included in the sampling during the initial screening.
Study design and sample size determination
The design of the study was cross-sectional and serum samples were collected from Foot-and- mouth disease-infected regions. The sample size for the serosurvey was initially determined and NSP 3ABC ELISA was performed. The aim of this particular study was to determine the serotype of the NSP3ABC ELISA positive sera using the Solid phase Competition ELISA. For this, representative sera samples with low OD values with NSP ELISA were selected and subjected to Solid phase competition ELISA. For this particular analysis 185 sera samples from cattle (Amhara = 99, Oromia = 34, Addis Ababa = 22 and Tigray = 30) were used.
Ethics approval and consent to participate
The study obtained research ethical clearance approved by the institutional animal research ethics committee at the Addis Ababa University, College of veterinary medicine and agriculture (Certificates Reference number: VM/ERC/18/03/13/2020 and the approval Date is 11/03/2020. All the methods were performed according to the guideline and regulations set by the university.
Laboratory analysis
Solid phase competition ELISA (Screening protocol): All serum samples collected from those mentioned regions were screened for the presence of NSP against Foot-and-mouth disease infection first. For this purpose, a total of 1421 serum samples were analyzed and found that 357 sera-positive animals were. A commercially available test kit (PrioCHECK® FMDV NSP, Netherlands) was used. This test is having specificity and sensitivity of 95% and 98% respectively according to the manufacturer’s instructions. Test plates of the kit contain FMDV NSP captured by the coated 3ABC specific mAb. This test is designed in such a way that it can identify antibodies of natural infection from the vaccinal antibody. The assay was performed according to the manufacturer’s instructions/protocol. The results were analyzed and interpreted using the Percentage Inhibition (PI) value of each sample. This test was validated by the manufacturer and marketed to be used as a diagnostic kit that identifies infected from vaccinated.
Serotyping using solid-phase competition ELISA: Those samples with low OD value (positive for 3ABC FMD) were selected and subjected to SPCE (solid phase competitive). The uniqueness of this study is that serotypes are identified from the serum of infected animals or positive 3ABC ELISA samples.
Principles of the solid-phase competition ELISA: ELISA microplates are supplied pre-coated with the FMDV serotypes inactivated antigens captured by the homologous MAb. Approximately diluted test sera are incubated with the trapped inactivated antigen, enabling the specific antibodies eventually present in the sample to bind to the respective antigen. Then, the anti-FMDV serotypes MAb, conjugated with peroxidase, are dispensed: its reaction with the homologous antigen will be inhibited by antibodies of positive sera previously bound to the virus, while in the case of negative sera the conjugated MAb can bind to the FMDV inactivated antigen. After incubation, the unbound conjugate is removed by washing and the TMB substrate/chromogen is delivered into wells. A calorimetric reaction develops if the conjugated MAb has bound to the virus, i.e. if test serum is negative, while color development is inhibited by positive sera. Inactivated antigen and conjugate concentrations are pre-calibrated to give a suitable reaction value (OD). After the addition of a stop solution, the optical density of the developed color is read by a microplate photometer at 459 nm.
Interpretation: The percentage inhibition produced by the positive control and by the test sera is calculated as follows:
% inhibition OD = 100-(serum OD/reference OD) ×100
Reference OD = mean OD of four wells processed with the Negative Control Screening test – test sera are considered:
- Positive when producing an inhibition ≥70% at the 1/10 dilution.
- Negative when producing an inhibition <70% at the 1/10 dilution.
Data analysis
Microsoft Excel spreadsheet was used to record and code the data generated from laboratory investigation and analyzed using the STATA version14 [8]. Sero-prevalence and stereotyping were analyzed by descriptive statistics.
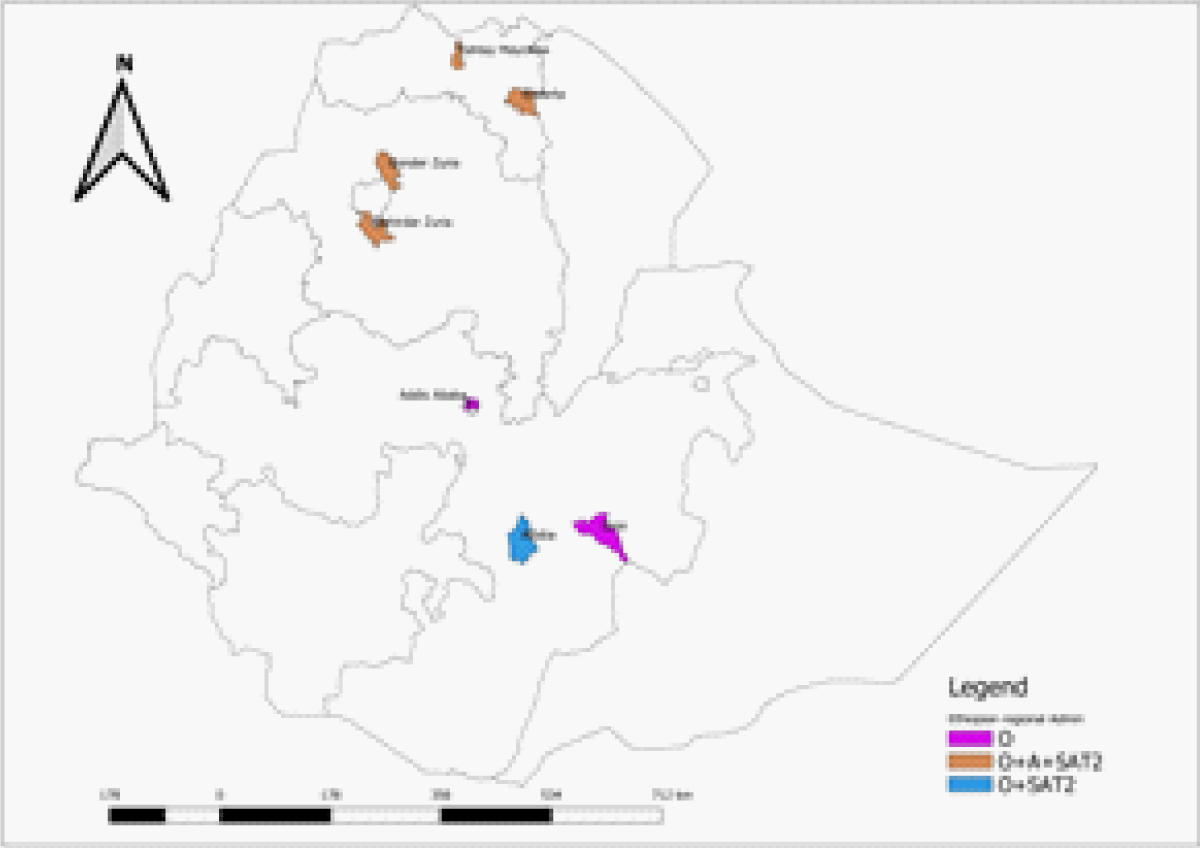
In this particular study, a total of 186 FMDV naturally infected animals with positive serum were analyzed for identifying FMDV antibody serotypes that are representing four regions of the country. The geographic distribution of the FMDV was indicated in the following Map where all three serotypes were found from the central to Northern part of the country. The northern part of the country harbors all three serotypes (Figure 1).
Figure 1: Geographic distribution of the FMDV antibody serotypes identified.
From 300 serum samples collected from Addis Ababa city, 53 serum samples were found to have antibodies against the non-structural protein. Twenty-three serum samples were further subjected to Solid phase competitive ELISA and serotype O was found in 10 of the samples, serotype A in 3 samples, and serotype SAT2 in 5 of the samples. 300 serum samples collected from the Oromia region were also analyzed and found that 51 samples were positive for FMDV NSP protein. Of those positive serum samples, 34 were subjected to the SPCE test and found that 12 were serotype O and 3 were serotype SAT2. Similarly, 435 serum samples originating from the Amhara region were tested with FMDV NSP ELISA and found that 104 of them were positive for natural infection antibodies. Of these 99 of them were analyzed using SPCE and all of them were serotype O and mixed infection was found in 15 of them with serotype A and SAT2 in 7 of the serum samples. From 386 serum samples collected from the Tigray region, 149 were seen to have an antibody for 3ABC FMDV NSP and all of them were seen to have Serotype O infection by the solid phase competition ELISA, in addition, 25 and 13 of them showed mixed infection with A and SAT2 serotypes (Table 1).
| Table 1: Regional distribution of antibody serotyping of 3ABC FMD positive serum samples. | ||||||
| Location | No of Sample | 3ABC ELISA positive |
Number of tested | SPCE | ||
| O | A | SAT2 | ||||
| Addis Ababa | 300 | 53 | 23 | 10 | 3 | 5 |
| Oromia | 300 | 51 | 34 | 12 | 0 | 3 |
| Amhara | 435 | 104 | 99 | 99 | 15 | 7 |
| Tigray | 386 | 149 | 30 | 30 | 25 | 13 |
The serotype distribution was also analyzed based on specific locations and found that serotype O was identified in all locations with the highest and lowest prevalence in Bahirdar and Ghinir, Adaba respectively. On the other hand serotype A and SAT2 were identified from the Mekelle area and Bahirdar with the highest proportion (Table 2). This serotype distribution was statistically significant at p < 0.05.
| Table 2: Serotype distribution of FMDV in different areas of Ethiopia. | ||||||
| Serotype | Bahirdar | Gonder | Mekelle | Addis Ababa | Ghinir | Adaba |
| O | 72 | 27 | 30 | 10 | 6 | 6 |
| A | 15 | 0 | 25 | 3 | 0 | 0 |
| SAT2 | 9 | 0 | 13 | 5 | 0 | 3 |
The proportion of serotypes identified based on the production system practices was also found that semi-intensive production takes the largest share in all three serotypes followed by intensive production (Table 3). There was a statistically significant association between the serotype identified and production system practices (p < 0.05).
| Table 3: Serotype distribution of FMDV with respect to the production system. | |||
| Serotype | intensive | Semi-intensive | extensive |
| O | 21 | 108 | 22 |
| A | 12 | 23 | 8 |
| SAT2 | 10 | 13 | 7 |
Past infection of the foot and mouth disease was confirmed by the presence of NSP antibodies which stay longer than Structural Protein (SP). In addition, studies have shown that antibodies against the non-structural protein (NSP) can persist for a longer duration than SP after infection [9]. Determination of serotype of foot-and-mouth disease virus has been done using antigen ELISA, Serotyping PCR and Solid-phase competitive ELISA.
In the present study three serotypes were identified from 186 NSP ELISA positive sera of which 156 serotype O, 40 serotypes A and 28 serotypes SAT2, this agrees with the reports of [4,10]. Another report by [10] also indicated that these three serotypes are circulating in the country.
In the current study serotype O was the most circulating serotype in all four regions, which agreed with previous studies [10,11] that reported serotype O was the dominant serotype causing outbreaks in Ethiopia Other studies also indicated that serotype O was the most common cause of outbreaks globally followed by serotype A [12,13]. These frequent occurrences of O serotype in different outbreak might be due to the presence of uncontrolled animal movement in the country. Additionally, this was in line with the findings of [4,14-16] who reported serotype O, A and SAT2 prevalence in order of dominancy.
The proportion of serotypes identified based on the production system practices was also found that semi-intensive production takes the largest share in all three serotypes followed by intensive production. There was a statistically significant association between the serotype identified and production system practices (p < 0.05).
Foot-and-mouth disease is one of the most contagious diseases in the world resulting in a highly devastating economic impact mainly in developing countries. Early diagnosis and typing of the agent are very important to effectively design and implement the control approach. In this particular study, it was found that serotypes O, A and SAT2 were identified by serotyping from previously infected animal targeting antibodies produced against the structural protein of the virus. Contrary to the Antigen ELISA (sandwich ELISA) and serotype-specific PCR which needs the antigen for the determination of the serotype, this one is found to be an alternative diagnostic technique with the comparative advantage which does not need the antigen (fear of escape of antigen) as well as no need of active outbreak. Early determination of the serotype from past infection helps to be aware of the epidemiology as well as the infection immunity of the herd/individual. This helps the design and implementation of the best control approach. Based on this the following recommendations are forwarded:-
➢ Large-scale sero-surveillance should be coupled with serotype identification from serum found positive for FMD past infection
➢ Previous infection immunity needs to be considered while designing a control approach
The authors would like to acknowledge AHI (former NAHDIC) and AAU, CVMA. In addition, all the farmers allowed us their animals for sampling.
Authors’ contributions
AM conceived and designed the study, Laboratory analysis FT: responsible for data integrity, analysis and interpretation, drafted and revised the manuscript. Both authors read and approved the final manuscript.
Funding
This research was supported by the Addis Ababa University (AAU) research and technology transfer office and AHI (former NAHDIC).
Availability of data and materials: The data used to support this study are available from the corresponding author on request.
Ethics approval and consent to participate: The study obtained research ethical clearance approved by the institutional animal research ethics committee at the Addis Ababa University, College of veterinary medicine and agriculture (Certificate Reference number: VM/ERC/18/03/13/2020, and the approval Date is 11/03/2020. All the methods were performed according to the guideline and regulations set by the university.
Informed consent statement: During serum sampling, the farmers were told about the research and the benefit afterward for their oral informed consent.
Conflicts of interest: The authors declare no conflict of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript, or in the decision to publish the results.
- Grubman MJ, Baxt B. Foot-and-mouth disease. Clin Microbiol Rev. 2004 Apr;17(2):465-93. doi: 10.1128/CMR.17.2.465-493.2004. PMID: 15084510; PMCID: PMC387408.
- Domingo E, Mateu MG, Martinez MA, Dopazo J, Moya A, Sorbino F. Genetic variability and antigenic diversity of foot and mouth disease virus. Appl Virol Res. 1990;2: 233–266. Available: https://www.cabdirect.org/cabdirect/abstract/19912218674
- Knight-Jones TJ, Robinson L, Charleston B, Rodriguez LL, Gay CG, Sumption KJ, Vosloo W. Global Foot-and-Mouth Disease Research Update and Gap Analysis: 1 - Overview of Global Status and Research Needs. Transbound Emerg Dis. 2016 Jun;63 Suppl 1:3-13. doi: 10.1111/tbed.12528. PMID: 27320162.
- Ayelet G, Soressa M, Sisay T, Belay A, Gelaye E, Jembere S, Skjerve E, Asmare K. FMD virus isolates: the candidate strains for polyvalent vaccine development in Ethiopia. Acta Trop. 2013 Jun;126(3):244-8. doi: 10.1016/j.actatropica.2013.02.005. Epub 2013 Feb 12. PMID: 23416124.
- Tekleghiorghis T, Moormann RJ, Weerdmeester K, Dekker A. Foot-and-mouth Disease Transmission in Africa: Implications for Control, a Review. Transbound Emerg Dis. 2016 Apr;63(2):136-51. doi: 10.1111/tbed.12248. Epub 2014 Jul 23. PMID: 25052411.
- Aman E, Molla W, Gebreegizabher Z, Jemberu WT. Spatial and temporal distribution of foot and mouth disease outbreaks in Amhara region of Ethiopia in the period 1999 to 2016. BMC Vet Res. 2020 Jun 9;16(1):185. doi: 10.1186/s12917-020-02411-6. PMID: 32517750; PMCID: PMC7285603.
- Callahan JD, Brown F, Osorio FA, Sur JH, Kramer E, Long GW, Lubroth J, Ellis SJ, Shoulars KS, Gaffney KL, Rock DL, Nelson WM. Use of a portable real-time reverse transcriptase-polymerase chain reaction assay for rapid detection of foot-and-mouth disease virus. J Am Vet Med Assoc. 2002 Jun 1;220(11):1636-42. doi: 10.2460/javma.2002.220.1636. PMID: 12051502.
- StataCorp. Stata Statistical Software: Release 14. College Station, TX: StataCorp LP. 2015. https:8//www.google.com/search?q=StataCorp.+2015.+Stata+Statistical+Software%3A+Release+14.+College+Station%2C+TX%3A+StataCorp+LP.&source=hp&ei=uwFwYMuSGtGJlwT2066wBQ&iflsig (accessed on 12 July 2022)
- Mohapatra JK, Priyadarshini P, Pandey L, Subramaniam S, Hemadri D, Sanyal A, Pattnaik B. Phylogenetic analysis of 3C protease (3C(pro)) coding region of Foot-and-mouth disease virus type A. Acta Virol. 2009;53(3):175-83. doi: 10.4149/av_2009_03_175. PMID: 19941399.
- Ayelet G, Mahapatra M, Gelaye E, Egziabher BG, Rufeal T, Sahle M, Ferris NP, Wadsworth J, Hutchings GH, Knowles NJ. Genetic characterization of foot-and-mouth disease viruses, Ethiopia, 1981-2007. Emerg Infect Dis. 2009 Sep;15(9):1409-17. doi: 10.3201/eid1509.090091. PMID: 19788808; PMCID: PMC2819860.
- Negusssie H, Kyule MN, Yami M, Ayelet G, Jenberie S. Outbreak investigations and genetic characterization of foot-and-mouth disease virus in Ethiopia in 2008/2009. Trop Anim Health Prod. 2011 Jan;43(1):235-43. doi: 10.1007/s11250-010-9683-2. Epub 2010 Aug 18. PMID: 20717724.
- Klein M, Hadaschik D, Zimmermann H, Eggers HJ, Nelsen-Salz B. The picornavirus replication inhibitors HBB and guanidine in the echovirus-9 system: the significance of viral protein 2C. J Gen Virol. 2000 Apr;81(Pt 4):895-901. doi: 10.1099/0022-1317-81-4-895. PMID: 10725414.
- Rweyemamu MM. Foot and mouth disease control strategies in Africa. Prev Vet Med. 1984;2: 329–340. doi:https://doi.org/10.1016/0167-5877(84)90076-X
- Sulayeman M, Dawo F, Mammo B, Gizaw D, Shegu D. Isolation, molecular characterization and sero-prevalence study of foot-and-mouth disease virus circulating in central Ethiopia. BMC Vet Res. 2018 Mar 27;14(1):110. doi: 10.1186/s12917-018-1429-9. PMID: 29587741; PMCID: PMC5870258.
- Sahle M, Venter EH, Dwarka RM, Vosloo W. Molecular epidemiology of serotype O foot-and-mouth disease virus isolated from cattle in Ethiopia between 1979-2001. Onderstepoort J Vet Res. 2004 Jun;71(2):129-38. doi: 10.4102/ojvr.v71i2.275. PMID: 15373335.
- Tesfaye Y, Khan F, Yami M, Wadsworth J, Knowles NJ, King DP, Gelaye E. A vaccine-matching assessment of different genetic variants of serotype O foot-and-mouth disease virus isolated in Ethiopia between 2011 and 2014. Arch Virol. 2020; 165(8):1749-1757. doi: 10.1007/s00705-020-04662-y. Epub 2020 May 20. PMID: 32435857.